The Air You're Breathing? A Diatom Made That
Andrew Alverson is a biologist and expert on diatoms at the University of Arkansas in Fayetteville. He contributed this article to Live Science's Expert Voices: Op-Ed & Insights.
You'll consume about 2 liters (just over a half gallon) of oxygen in the time it takes you to read this post. About 20 percent of that oxygen comes from photosynthesis by marine diatoms — the most important little organisms that most people have never heard of.
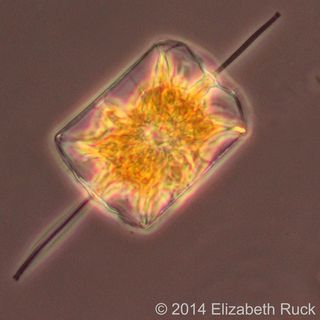
Diatoms are tiny — five to 10 of them could fit on the head of a pin — but these single-celled algae play an immense role in keeping the planet's ecosystem working. They're important mediators of carbon and oxygen cycles, an integral component of marine food webs and the principal cyclers of silica, which constitutes about one-quarter of the Earth's crust. [In Photos: The Diversity of Diatoms]
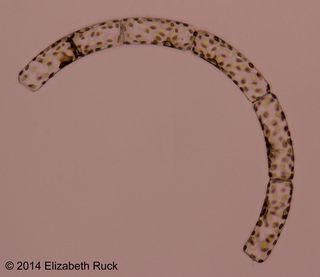
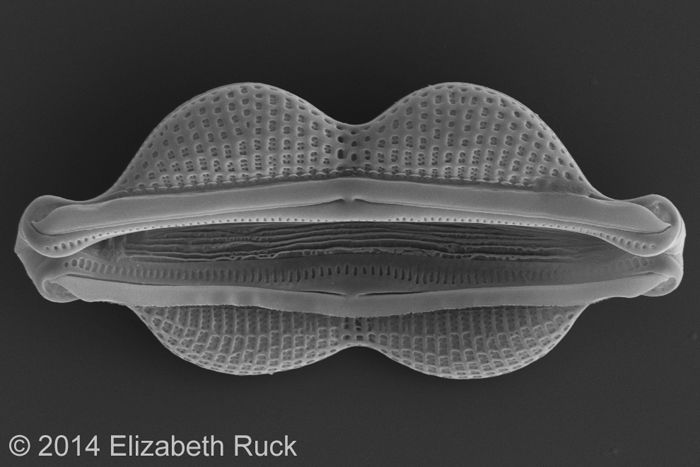
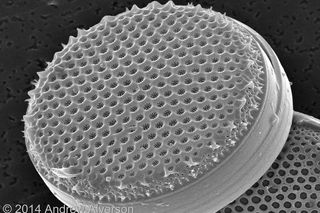
Diatoms incorporate that silica into their beautifully ornamented glass cell walls, whose intricate patterns have captivated researchers for centuries. Diatom species are distinguished largely on the basis of their cell-wall features and, increasingly, differences in their DNA sequences. No one really knows how many different diatoms are out there, but conservative estimates suggest around 100,000 to 200,000 species, making them among the most species-rich lineages of eukaryotes. Eukaryotes include plants, animals, and everything else whose cells are organized into membrane-bound compartments, including a cell nucleus.
Found in environments ranging from tropical reefs to sea ice , and freshwater to very salty waters — and just about every place in between — diatoms are ecologically and physiologically diverse as well. All of this has motivated a great deal of research on identifying the factors that led to their extraordinary morphological, ecological and species-level diversity.
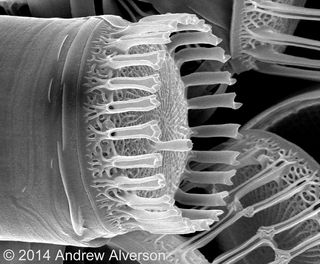
Advances in DNA sequencing technologies have made it possible, and practical, to address these types of questions with whole-genome sequencing. Just a few diatom genomes have been sequenced so far, but the results suggest that their genomes are likely to hold many clues to their origin and diversification — their evolutionary "success."
Diatoms appear to have a highly mosaic genome, with genes originating from many different sources. Most notably, a large fraction of the genes may have been acquired by horizontal gene transfer (HGT) from bacteria. Although genomic data have shown that HGT — the swapping of genes between species that don't reproduce with one another — is much more common in eukaryotes than once thought, gene transfer between such distant relatives (diatoms and bacteria last shared a common ancestor a few billion years ago) is rare.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.

Determining that a diatom gene was acquired from bacteria is not as easy as it might seem, and the determination is likely to vary — either subtly or dramatically — depending on how many sequenced genomes are available for comparison. Much in the same way that police can't find a perpetrator if his fingerprints aren't in their database, it would be difficult, if not impossible, to figure out that a gene resembling one from bacteria is an ancient, established eukaryotic gene if there are no eukaryotic genomes to compare against and analyze.
Given that hundreds of eukaryotic genomes have been sequenced since the diatom-bacterial HGT discovery in 2008, a much more refined story is likely to emerge in the coming years. In particular, projects funded by the Gordon and Betty Moore Foundation and the U.S. National Science Foundation will produce complete genomes for several more diatoms and transcriptomes (most of the organism's genes) for hundreds of additional diatom species. Those data will shed valuable new light on the extent to which diatoms have benefited from extrinsically derived genetic material.
It remains to be seen whether some of the same, mostly intrinsic, processes implicated in the diversification of other "hyperdiverse" eukaryotes have also been at play over the course of diatom evolution. Flowering plants and vertebrates, for example, famously experienced serial rounds of whole-genome duplication. Although most of the duplicated genes were almost immediately lost into the dustbin of evolutionary history, some of the duplicates remained, freely evolving independently of their progenitors to acquire new or modified functions. Diatom genomes contain lots of duplicated genes, but whether these originated piecemeal or in large, punctuated bursts is an open question.
So, what makes a diatom a diatom? What factors helped pave the way to their current place of prominence in the world's oceans? What causes some lineages, like diatoms, to diversify while others languish? Can genes and genomes help us answer these questions?
Like land plants and other eukaryotic algae, diatom plastids — the engines of photosynthesis — trace back to once free-living bacteria. But do diatoms owe a special debt to bacteria? To what extent have adopted bacterial genes been a source of important new traits in diatoms? To what extent has diatom diversification followed the same routes as other diverse, but more intensively studied, eukaryotes? With a recent grant from the National Science Foundation, a colleague — Norm Wickett from the Chicago Botanic Garden — and I will use, and contribute to, the coming surge of genomic data to help answer some of these questions.
Genomes won't fill all of the gaps, however. By most estimates, some 90 percent of diatom species have yet to be discovered and named, so even as we begin to unravel the story of diatom diversification, the vast majority of the players will remain anonymous.
Editor's Note: You can here a public radio interview with Alverson on KUAF.
Follow all of the Expert Voices issues and debates — and become part of the discussion — on Facebook, Twitter and Google +. The views expressed are those of the author and do not necessarily reflect the views of the publisher, or the University of Arkansas. This version of the article was originally published on Live Science.